Radiologia Brasileira - Publicação Científica Oficial do Colégio Brasileiro de Radiologia
AMB - Associação Médica Brasileira CNA - Comissão Nacional de Acreditação
 Vol. 40 nº 4 - July / Aug. of 2007
Vol. 40 nº 4 - July / Aug. of 2007
|
ORIGINAL ARTICLE
|
|
Grid computing in the optimization of content-based medical images retrieval |
|
|
Autho(rs): Marcelo Costa Oliveira, Paulo Mazzoncini de Azevedo-Marques, Walfredo da Costa Cirne Filho |
|
|
Keywords: Content-based image retrieval, Texture analysis, Image registration, Grid computing |
|
|
Abstract:
IPhD of Medicine in the field of Medical Information Technology, Master in Physics applied to Medicine, Bachelor in Computing Sciences, Professor at Universidade Federal de Alagoas, Campus Arapiraca, Arapiraca, AL, Brazil
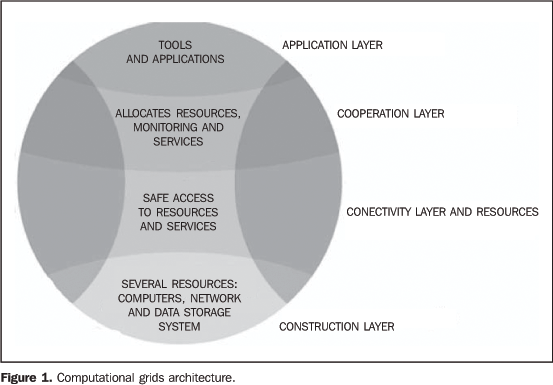
INTRODUCTION The amount of data generated in hospitals and medical centersgrows at increasing rates. The yearly production of datagenerated by diagnostic imaging in important radiological centersachieves two terabytes, as a result of acquisition and storage ofdata regarding patients in consequence of the increasingimportance and utilization of imaging diagnosis. There is a needfor intelligent and safe indexation and storage of a huge amountof data, considering that they play an essential role in clinicaldiagnosis(1,2). The increasing use of computer-aided diagnosis (CAD)applications is related to the rapid development of medicalalgorithms. The CAD objective is to improve the diagnosticaccuracy, as well as the consistency in diagnostic imagesinterpretation by means of a diagnostic response suggested by acomputer(3). However, some recognized CADapplications have not been utilized in the clinical routine yetbecause of their high computational cost, limiting theirutilization to those centers that have high capacitycomputers(4). Difficulties in applying these CAD algorithms in the clinicalroutine and the current limits on images storage, processing,search and retrieval in large data banks have led companies andresearch institutions to find new solutions for theaccomplishment of these tasks(5,6). Grid computing The grid computing (GC) technology is the most recent andpromising tool in the field of distributed computing. In summary,distributed computing consists of a collection of independentcomputers presenting to the user as a single and consistentsystem. This technology allows integration between remotelydistributed and connected by means of long distance networks.With this integration capacity, a virtual or cooperativecomputing system is created to resolve problems regarding massdata storage and access, as well as regarding applicationsrunning with high computational cost(6,7). The GC technology offers a single environment for datasharing, storage and processing. Additionally, it allows themedical community to utilize a single distributed databasecapable of providing resources, data and knowledge sharing. Thesecapabilities allow a greater interaction among medical clinics,offering new opportunities for small clinics and researchlaboratories with poor computationalresources(7). Besides, GC offers a flexible, scalable and less vulnerableinfrastructure, and, therefore more reliable and capable ofguaranteeing a safe access to any installed application.Differently from distributed computing and the methodology ofgroups of terminals or workstations connected to a server orgroup of servers (clusters technology), the GC resources presentadministrative autonomy and system heterogeneity. These twofeatures allow a higher scalability and robustness toapplications. However, these same features require that thecomputational grid components are compliant with standards aimingat an open scalability and sharing of computationalresources(8). Foster & Kesselman(9) have presented a proposal of GC architecture and components. The grid formal architecture comprises four layers (Figure 1). The construction layer is the lowest level of the structure and represents the physical resources and devices that users want to share and access (computers, network, file systems, catalogs, softwares and digital instruments). Just above the construction layer, is the resources and connectivity layer, responsible for communication and authentication required for resources exchange, user validation, monitoring and control over resources sharing. The third layer, or cooperation layer, holds the protocols and performs the services responsible for the resources exchange (resources discovery and allocation, monitoring and diagnosis of services functionality, data replication, and policies regulating users' privileges for accessing the grid resources). The user application layer is at the top of the structure and is responsible for invoking all the other layers.
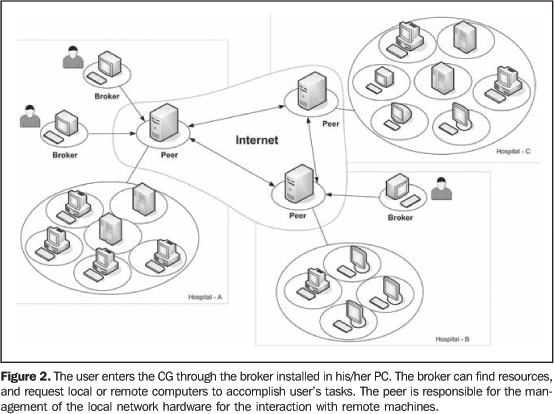
There is a great number of GC-related projects described inthe literature (for example: Globus(10),Legion(11), Condor(12) andOurGrid(13)), based on different technologies,and aimed at determined areas and purposes like applications fordata storage and processing, Web portals and infrastructureservices for interinstitutionalcollaboration(14,15). The baseline utilization of GC by the user is accomplished by means of a software interface that allows the user to communicate with the data processing center of the computational grid known as broker. The broker can find the resources required for the tasks execution. After finishing the task execution, the broker returns the application result to the user(16). Figure 2 demonstrates the baseline functioning of the OurGrid project.
Content-based medical images retrieval Among the several CAD techniques, content-based imagesretrieval (CBIR) are the systems that most benefit from the GCtechnology due to their features and requirements: processingintensity and complexity and great amount of storedimages(17). Through CBIR, and based on a reference image, it is possibleto find similar images included in one or several image banksutilizing inherent attributes. In the clinical decision-makingprocess, CBIR presents great advantages, and is capable ofretrieving images of a same modality, anatomical region and withthe same structural alterations caused by certain diseases.Therefore, CBIR has awakened the medical community interestbecause of its capacity to retrieve already diagnosed images tocompare with an image being studied, allowing the specialist toconfirm his/her diagnostic hypothesis(18).Although part of this information may be shown on the medicalimages letterhead, this textual labeling may present a high rateof error, with case reports of up to 16%(19). Agreat number of scientific papers emphasize the need for adoptingalternative methods of accessing data manually inserted into themedical images letterheads(20–23). Besides the techniques of clinical decision-making support,research and teaching benefit from CBIR systems. In education,CBIR aids both teachers and students in utilizing educationalimage banks and visual analysis of results. Besides evaluationbased on diagnosis and anatomical region, analysis of visuallysimilar cases, although with different diagnosis, result in animprovement of the educational quality(24). Content-based images retrieval is one of the computationalvision techniques more intensely studied in the last tem years,and is based on three classes of visual characteristics: color,texture and shape(25). These attributes allowthe development of robust computational tools capable ofcharacterizing images by their own contents, adding advantages tothe images identification based only on textual descriptors thatconstitute the traditional classification of medical imagesfiles(23). The gray-scale distribution is the simplest feature to becharacterized. Characterization is performed by comparisonbetween gray-scale histograms utilizing the summation of absoluteor quadratic differences on the number of image elements (pixels)for each gray-scale intensity. Gray-scale distribution presentsambiguity (where different images may generate the samesummation), and for this reason is not effective for the wholeCBIR; however, considering its simplicity and low computationalcost it can and should be utilized as a initial filter for othermore complex and costly methods. Texture-based features are related to the quantification ofimage intensity variation and scale. In the literature, one ofthe most frequently utilized methods for extracting textureattributes is the co-occurrence matrix(24).Haralick et al.(26) have defined the textureattributes that can be obtained from the co-occurrence matrixwith texture discrimination purposes. Approximately 20statistical functions are proposed in the literature foracquisition of information from the co-occurrencematrix(27). Some of the most significantfunctions producing a satisfactory textures classification are:entropy, inertia, energy, shade, inverse difference moment,promenance, correlation andvariance(27–33). Shape-based image retrieval is one of the most complex issuesto be approached by CBIR systems, considering the complexity ofthe method for automatic segmentation of medical images. Afterthe segmentation, the structures are described by their shapecharacteristics, including information on rotation, translationand scale(34). Another CBIR technique described in the literature is theimages registration(5,17). This techniquecalculates a rigid 2D coordinates transformation includingrotation, translation and scale, searching the maximum matchingbetween two images or between two image volumes. The rigidtransformation is based on the minimization of the quadraticerror or sum of square differences between the structures contourutilizing similarity measurement algorithms between two imagesintensities(35,36). The present study presents a singular approach to systems ofcontent-based medical images retrieval, utilizing textureattributes and the computational power of the recent GCtechnology applied to the similarity measurement algorithm basedon the sum of square differences.
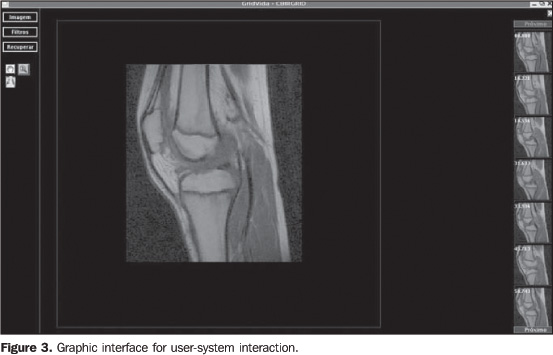
MATERIALS AND METHODS The system developed in the present study has utilized theGNU/Linux Debian operational system and Java 1.5 programminglanguage, with the similarity measurement algorithm as animplementation of the Insight Toolkitsoftware(37). Evaluation was made in aheterogeneous images bank with 2,400 MRI images of differentanatomical regions, sequences and acquisition planes, with graylevels ranging from 4,096 to 65,536. The system comprises two CBIR modules. The first moduleutilizes second order texture analysis parameters (co-occurrencematrix) to classify the most similar images according to thistechnique. In the second module, the similarity measurementalgorithm is applied on the images selected in the first module.Because of the high computational cost of the similaritymeasurement algorithm, the second module is processed on theOurGrid computational grid that is a cooperative, open andfree-access network. OurGrid, currently, hooks togetherapproximately 500 machines. The user/GC interface is performed by means of MyGrid 3.2(OurGrid; Campina Grande, PB, Brazil) that is the OurGrid broker,capable of selecting the computational resources to be utilizedin the application execution, besides releasing the user from theGC complexity, so the user utilizes de grid as if it was a singlecomputer(13). All of the database images have an associated characteristicvector obtained from the gray levels co-occurrence matrix and itsattributes. The co-occurrence matrix followed orientation at0°, 45°, 90° and 135° and distances betweenimages elements (pixels) = 1. Texture attributes utilized were:energy, entropy, inverse difference moment, shadow, inertia,promenance, correlation and variance. The utilization of eighttexture attributes and four angular orientations resulted in a32-dimension characteristic vector. The system offers a graphic interface (Figure 3) allowing the specialist to select a DICOM (digital imaging and communication in medicine) reference image at the beginning of the first module. At the end of the module, the images are classified according the lower value of the Euclidean distance between the characteristic vectors of the reference image and the database images.
The second module utilizes the 1,000 most similar imagesaccording to the first module. This module also requires that thespecialist define the number of tasks for the similaritymeasurement algorithm processing e distribution on the GC. Thatis to say, which is the application "granularity". Thegranularity is related to the amount of images to be processed bythe similarity measurement algorithm on each GC machine. Thesimilarity measurement algorithm utilizes similar transformationsand linear interpolation aiming at the mapping of the homologuepoints between two images.
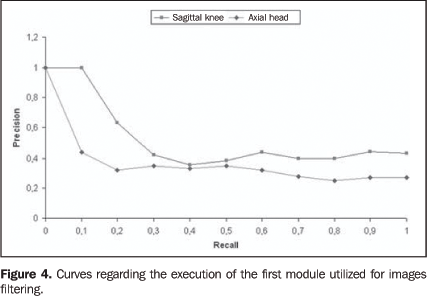
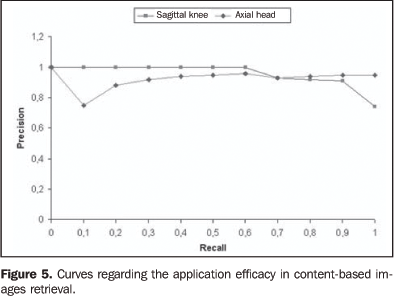
RESULTS The results of the present study originated from the selectionof images of two anatomical regions — knee and head —in an images bank. The knee studies included 20 sagittal,T1-weighted images, and head studies included 40 axial,T2-weighted images. The experiments were repeated for threetimes, with slices different from the described studies. Theimages were considered as correct when the application returnedimages from the same plane and sequence of the referenceimages. The first module classified the most similar images accordingto texture attributes. The mean processing time in the firstmodule was 2.3 minutes, and was obtained by the calculation ofthe Euclidean distance between the characteristic vector of eachof the 2,400 images, and the characteristic vector of thereference image. The algorithms were processed by the localcomputer utilizing a 2.8 GHz Pentium 4 processor with 1 Gbytememory. Results were evaluated with "precision" and "recall"parameters which are typically utilized for evaluating systems ofcontent-based images retrieval and information retrieval."Recall" means the ratio of relevant images over the number ofimages retrieved in the query. On the other hand, "precision" isthe ratio of retrieved images that are relevant forreference(38). Figure 4 shows the results of the execution of the first module with mean values of precision-recall curves of the Euclidean distance between characteristic vectors of the reference images in relation to the images of the database. This result allowed the evaluation of the CBIR effectiveness utilizing the texture in the classification of the most similar images for the second module. Although the mean precision obtained in the experiments is 0.54 (sagittal knee), and 0.40 (axial head), it is sufficient for filtering the images to be submitted to the second module. In the second module, the images are processed with the similarity measurement algorithm on the computational grid. CBIR with the similarity measurement algorithm resulted in a satisfactory precision for both anatomical regions — 0.95 (sagittal knee) and 0.92 (axial head) —, according to the mean precision-recall curves between the reference images and those classified by the first module (Figure 5).
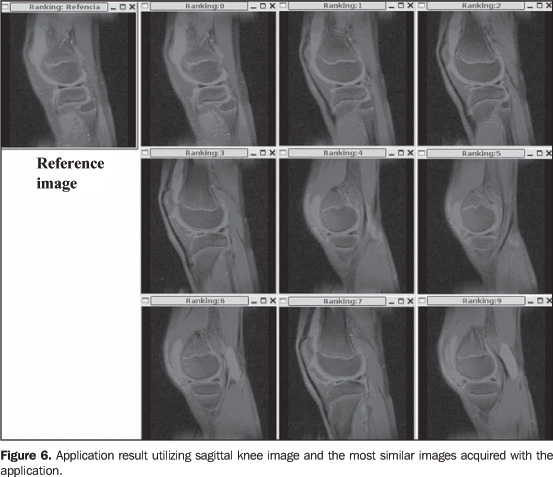
Figure 6 shows the classification of the most similar images after the application execution. For space reasons, only nine of the most similar images are shown.
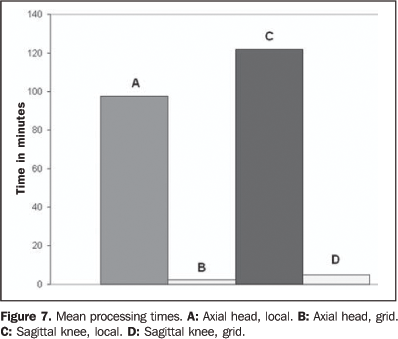
The high computational cost of the similarity measurement algorithm was balanced by the utilization of the computation grid of the OurGrid system. On average, the processing time of the similarity measurement algorithm applied to the experiments utilizing 50 processor of the grid was reduced by 116.97 minutes for knee images and 95.15 minutes for head images in relation to processing times obtained in the local computer (Figure 7).
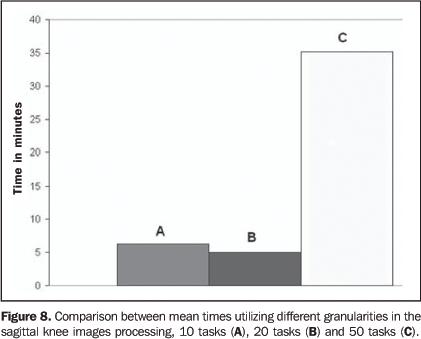
In the present study, the application was divided into 20tasks comprised of 50 images each. Images were compressed beforebeing sent to the computational grid, and the mean size of thefiles with 50 images was 4 Mbytes. Images were sent to thecomputational grid with a single identification file specifyingthe number of the image and the respective task. On average, the compressed images were sent to thecomputational grid machines is 22.2 seconds, and the meanprocessing time for the 50 images by each computer of the gridwas 11.45 seconds. The images send-time was short because thegreatest part of the tasks was executed on computers connected tothe local network. Also, OurGrid allowed that the libraries required for theapplication execution were stored in remote computers avoidingthe necessity of re-sending data. The mean time for experiments has also been analyzed, changing the application granularity among 10, 20, and 50 images/task (Figure 8). The use of the smallest grain, i.e., 50 images /task, implied a greater total amount of images/task. So, a higher number of computers of the computational grid were requested because of the increase in the quantity of tasks to be processed.
The necessity of allocating 50 machines for executing theapplication implied the distribution of tasks for being processedout of the local network. So, the total application time wasaffected by the time of data transmission to remotecomputers. Nevertheless, decomposing the application into larger tasks(tem tasks in total), i.e., larger grain, implied the requisitionof less computers and transmission of greater files with higherprocessing time/machine. Therefore, a fixed and intermediatenumber of 20 tasks were adopted.
DISCUSSION The CG technology has shown to be a promising tool in theprocessing and storage of great data volumes. However, morebenefits should be expected, according to Liu etal.(7), who have utilized the GC architectureto make medical images backup copies in several PACS (picturearchiving and communication system). The present study has adopted a mixed approach of CBIRtechniques to classify similar images of different planes andanatomical regions utilizing the high CG processing capacity. Thesystem has utilized CBIR techniques based on texture analysis andsimilarity measurement algorithm. The texture analysis approximates the human visual perceptionand has been utilized in many systems as an aid to the clinicaldiagnosis(39,40). The mean texture analysisaccuracy — 0.54 for knee images, and 0.40 for head images -,despite being relatively low, was effective as an initial filterfor the second module. A possible solution to increase theefficiency of this filtering would be the development of methodsto detect motion artifacts, since texture information may bemissed when rotation, translation and scaling are involved in theimages processing(29). The utilization of the similarity measurement algorithm of thesum of the square differences applied to the second modulepresented a quite satisfactory mean accuracy — 0.95 forknee, and 0.92 for head. The algorithm could retrieve similarimages of different anatomical regions and planes. The majorityof studies in the literature are restricted to a determinedanatomical region, modality or diagnostic procedure, onlyutilizing characteristic vectors(41). However,because of their high computational cost, the utilization ofsimilarity measurement algorithms executed in a single computerbecomes unfeasible in computer-aided diagnosis. The GC technologyenables the utilization of the similarity measurement techniquebecause of the capability of parallel data processing in theseveral computers connected to the computational grid. Although the computational grid utilized in the present studyis constituted by approximately 500 computers spread over morethan 20 locations, the ten- and twenty-task experiments wereprocessed in the local network machines without affecting theapplication execution time. However, the 50-task experimentsrequired processing out of the local network, so they wereaffected by the high costs of data transmission. In these cases,the cost-benefit ratio between processing time anddata-transmission should be evaluated. The utilization of GC in medical applications is still at itsbeginning; however this is a promising technology and significantdevelopments in IT applied to the health care field can beexpected in the near future. Aiming at improving the results of the present study, two newcomponents are presently in development: similarity measurementbased on cross-correlation, and automatic segmentation of brainstructures. The cross-correlation algorithm will allow the searchin different modalities to minimize a limitation of the sum ofsquare differences. Another limitation of this algorithm is thehigh sensitivity to small amounts of pixels with greatdifferences in intensity between two images, like in cases ofcontrast injection(35). The automaticsegmentation algorithm will restrict the image retrieval todetermined structures, allowing more specific queries than thoseperformed in comparison with complete images. An integrateutilization of different methods could result in a more accuratedifferentiation between images(42). Acknowledgments The authors thank Projeto GridVida, Laboratório deSistemas Distribuídos da Universidade Federal de CampinaGrande (UFCG), and Centro de Ciências das Imagens eFísica Médica da Faculdade de Medicina deRibeirão Preto da Universidade de São Paulo(CCIFM/FMRP-USP).
REFERENCES 1. Montagnat J, Breton V, Magnin IE. Using technologies to face medical image analysis challenges. Proceedings of the IEEE CCGrid03 2003, Tokyo, Japan. [ ] 2. Montagnat J, Breton V, Magnin IE. Partitioning medical image databases for content-based queries on a Grid. Methods Inform Med 2005;44: 154–160. [ ] 3. Azevedo-Marques PM. Diagnóstico auxiliado por computador na radiologia. Radiol Bras 2001;34: 285–293. [ ] 4. HealthGrid, HealthGrid White Paper. [Acessado em: 10/10/2006]. Disponível em: http://www. heathgrid.org [ ] 5. Montagnat J, Bellet F, Benoit-Catin H, et al. Medical images simulation, storage, and processing on the European DataGrid testbed. J Grid Comput 2004;2:387–400. [ ] 6. Breton V, Blanchet C, Legré Y, Maigne L, Montagnat J. Grid technology for biomedical applications. Lecture Notes in Computed Science 2005; 204–218. [ ] 7. Liu BJ, Zhou MZ, Documet J. Utilizing data Grid architecture for the backup and recovery of clinical image data. Comput Med Imaging Graph 2005;29:95–102. [ ] 8. Foster I, Kesselman C, Tuecke S. The anatomy of the Grid: enabling scalable virtual organizations. International Journal of High Performance Computing Applications 2001;15:200–222. [ ] 9. Foster I, Kesselman C. The Grid 2: blueprint for a new computing infrastructure. San Francisco, CA: Morgan Kaufmann Publishers, 2004. [ ] 10. Foster I, Kesselman C. Globus: a metacomputing infrastructure toolkit. International Journal of Supercomputing Applications 1997;11:115–128. [ ] 11. Grimshaw AS, Wulf WA. The legion vision of a worldwide virtual computer. Communications of the ACM 1997;40:39–45. [ ] 12. Condor. [Acessado em: 13/9/2006]. Disponível em: http://www.cs.wisc.edu/condor [ ] 13. Cirne W, Brasileiro F, Andrade N, et al. Labs of the World, Unite!!! UFCG/DSC Technical Report 07/2005;1–12. [ ] 14. de Roure D, Baker M, Jennings NR, Shadbolt N. The evolution of the Grid. In: Berman F, Fox G, Hey AJG, editors. Grid computing – making the global infrastructure a reality. New York, NY: Wiley, 2003;65–100. [ ] 15. Foster I. The Grid: computing without bounds. Scientific American April 2003,228:80–85. [ ] 16. Grid Café. [Acessado em: 29/8/2006]. Disponível em: http://gridcafe.web.cern.ch/gridcafe [ ] 17. Montagnat J, Duque H, Pierson JM, Breton V, Brunie L, Magnin IE. Medical image content-based queries using the Grid. Proceedings of the First European HealthGrid Conference 2004, Lyon, France. [ ] 18. Rahman M, Wang T, Desai B. Medical image retrieval and registration: towards computer assisted diagnostic approach. In: IDEAS Workshop on Medical Information Systems: The Digital Hospital (IDEAS-DH'04), 2004. Washington, DC: IEEE Computer Society, 2004;78–89. [ ] 19. Güld MO, Kohnen M, Keysers D. Quality of DICOM header information for image categorization. Proceedings of the International Symposium on Medical Imaging 2002, San Diego, CA. [ ] 20. Tagare HD, Jaffe C, Duncan J. Medical image databases: a content-based retrieval approach. J Am Med Inform Assoc 1997;4:184–198. [ ] 21. Traina Júnior C, Traina AJM, Santos RR, Senzako EJ. A support system for content-based medical image retrieval in object oriented databases. J Med Syst 1997;21:339–352. [ ] 22. Rosset A, Ratib O, Valle J. Integration of a multimedia teaching and reference database in a PACS environment. RadioGraphics 2002;22:1567–1577. [ ] 23. Petrakis EGM. Content-based retrieval of medical images. Int J Comput Res 2002;11:171–182. [ ] 24. Müller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medical applications – clinical benefits and future directions. Int J Med Inform 2004;73: 1–23. [ ] 25. Azevedo-Marques P, Honda MH, Rodrigues JAH, et al. Recuperação de imagem baseada em conteúdo: uso de atributos de textura para caracterização de microcalcificações mamográficas. Radiol Bras 2002;35:93–98. [ ] 26. Haralick RM, Shanmuga K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern 1973;SMC3:610–621. [ ] 27. Walker RF, Jackway P, Longstaff ID. Improving co-occurrence matrix feature discrimination. In: Proc DICTA '95, 3rd Conference on Digital Image Computing: Techniques and Application 1995;643–648. [ ] 28. McLean GF. Vector quantization for texture classification. IEEE Trans Syst Cybern 1993;23:637–644. [ ] 29. Freeborough PA, Fox NC. MR image texture analysis applied to the diagnosis and tracking of Alzheimer's disease. IEEE Trans Med Imaging 1998;17:475–479. [ ] 30. Mathias JM, Tofts PS, Losseff NA. Texture analysis of spinal cord pathology in multiple sclerosis. Magn Reson Med 1999;42:929–935. [ ] 31. Materka A, Strzelecki M. Texture analysis methods – a review. In: COST B11 Report. Lodz, Poland: Technical University of Lodz, Institute of Electronics, 1998. [ ] 32. Konak ES. A content-based image retrieval system for texture and color queries. (M.Sc. degree thesis). Ankara, Turkey: Department of Computer Engineering and Institute of Engineering and Science, Bilkent University, 2002. [ ] 33. Sharma M, Singh S. Evaluation of texture methods for image analysis. 7th Australian and New Zealand Intelligent Information System Conference. Perth, Australia, 2001;117–121. [ ] 34. Veltkamp RC, Hagedoorn M. State-of-the-art in shape matching. In: Lew M, editor. Principles of visual information retrieval. London: Springer-Verlag, 2000;87–119. [ ] 35. Hajnal JV, Hill DLG, Hawkes DJ. Medical image registration. In: Neuman MR, editor. Biomedical engineering. Boca Raton, FL: CRC Press, 2001. [ ] 36. Yoo TS. Insight into images: principles and practice for segmentation, registration, and image analysis. Wellesley, MA: AK Peters, 2004. [ ] 37. InsightToolkit. [Acessado em: 8/10/2006]. Disponível em: http://www.itk.org [ ] 38. Bueno JM. Suporte à recuperação de imagens médicas baseada em conteúdo através de histogramas métricos. São Carlos, SP: Instituto de Ciências Matemáticas e de Computação, Universidade de São Paulo, 2002. [ ] 39. Shyu CR, Bradley CE, Kak AC, Kosaka A, Aisen AM, Broderick LS. ASSERT: a physician-in-the-loop content-based retrieval system for HRCT image databases. Computer Vision and Image Understanding 1999;75:111–132. [ ] 40. Kuo WJ, Chang RF, Lee CC, Moon WK, Chen DR. Retrieval technique for the diagnosis of solid breast tumors on sonogram. Ultrasound Med Biol 2002;28:903–909. [ ] 41. Lehmann TM, Güld MO, Thies O, et al. Content-based image retrieval in medical applications. Methods Inform Med 2004;43:354–361. [ ] 42. Traina AJM, Traina C, Bueno JM, Chino FJT, Azevedo-Marques P. Efficient content-based image retrieval through metric histograms. World Wide Web J 2003;6:157–185. [ ]
Received November 21, 2005. Accepted after revision October 23, 2006.
* Study developed in the Laboratório de Sistemas Distribuídos do Departamento de Sistemas e Computação da Universidade Federal de Campina Grande (UFCG), Campina Grande, PB, Brazil. |
|
Av. Paulista, 37 - 7° andar - Conj. 71 - CEP 01311-902 - São Paulo - SP - Brazil - Phone: (11) 3372-4544 - Fax: (11) 3372-4554