Radiologia Brasileira - Publicação Científica Oficial do Colégio Brasileiro de Radiologia
AMB - Associação Médica Brasileira CNA - Comissão Nacional de Acreditação
 Vol. 47 nº 5 - Sep. / Oct. of 2014
Vol. 47 nº 5 - Sep. / Oct. of 2014
|
ORIGINAL ARTICLE
|
|
Application for internal dosimetry using biokinetic distribution of photons based on nuclear medicine images |
|
|
Autho(rs): Viriato Leal Neto1; José Wilson Vieira2; Fernando Roberto de Andrade Lima3 |
|
|
Keywords: Nuclear medicine; Biokinetic distribution; Dosimetric evaluations; Administered activity. |
|
|
Abstract: INTRODUCTION
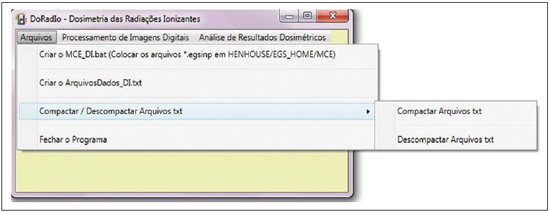
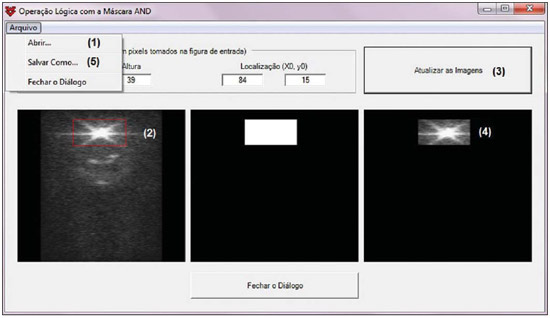
For diagnosis and therapy of various diseases, nuclear medicine uses radiopharmaceutical substances that have a radionuclide in their composition. The radiopharmaceutical physicochemical characteristics determine its biokinetics, i.e., its fixation in the target organ, metabolization and elimination, while the physical characteristics of the radio-nuclide determine the application of the compound in the diagnosis or therapy. Images from patients submitted to treatment with radioactive substances may be used to assess the distribution of the activity to be administered. The definition of amount of administered activity is generally under the responsibility of the nuclear physician who relies on pre-dose scanning of the whole body images of the patient (anteroposterior and posteroanterior views). Based on the presence of hot-spots on the images, the physician estimates the activity to be administered in the treatment. Several tasks must be accomplished to estimate the dose distribution in organs and tissues. Such tasks range from the phantom selection, its coupling with the Monte Carlo (MC) code and additional algorithms to simulate specific sources, to organization and presentation of tables and charts showing the absorbed dose accumulated in target organs selected as a function of different energy values of the photon. The present study discusses the development of the software called DoRadIo (Dosimetria das Radiações Ionizantes - stands for Dosimetry of Ionizing Radiations) aimed at integrating the above mentioned tasks into a single computer-aided tool. Menus clearly demonstrate such tasks for data obtention and organization into tables and charts for further analysis. The current version of the software is intended for the use by dosimetry investigators. With the dosimetry data available at the current version, it is possible to interpolate results since the source is a monoenergetic gamma emitter. In such a case, the user should save the table of the absorbed dose per accumulated activity for the problem situation of interest. The user should only to choose the source organ, the target and select the data on the table with results related to the energy of interest. The interpolation must be made with another software. Also, it is possible to obtain results when the photons are emitted from several source organs. The techniques that allow further dosimetry evaluations characterize the implemented innovations. The DoRadIo software is an integral part of Sistema de Informações Dosimétricas (SID) (stands for Dosimetric Information System)(1) developed and maintained by the Grupo de Pesquisa em Dosimetria Numérica (GDN/CNPq), whose website is http://dosimetrianumerica.org/. MATERIALS AND METHODS The software was built within the Microsoft Visual Studio 2010 with the project model WPF Application(2). Various tools were brought in from other GDN softwares, such as MonteCarlo(3) and Digital Image Processing (DIP)(4). As the MC simulations were restricted to gamma emitters, one resorted to EGSnrc (Electron Gamma Shower National Research Council)(5) code. The exposure computational model (ECM) MSTA (Mash STAnding)(6) provided by the Department of Nuclear Energy of Universidade Federal de Pernambuco (DEN-UFPE), Recife, PE, Brazil, (www.caldose.org) was inserted in the user interface system. The ECM MSTA consists of a voxel phantom representing a male adult body coupled with EGSnrc. The MSTA user code comprises 13 source simulators for external dosimetry and one for internal dosimetry, this particular one with 27 text files with data on sources recommended for medical practice. The MSTA compilation was performed at the EGSnrc user interface (UI), and the code ran by the Windows command prompt. All necessary softwares were installed in the computers (Intel Core i7 X990® 3,47 GHz processors, 24 GB RAM memory with the 64 bits Windows 7 Ultimate operational system) at Laboratório de Dosimetria Numérica (LDN) of the Instituto Federal de Educação, Ciência e Tecnologia de Pernambuco (IFPE). The methods incorporated into the menus Files (Arquivos) and Digital Images Processing (Processamento de Imagens Digitais) of the DoRadIo software are presented below, with details on the functionalities of each menu. The Files (Arquivos) menu The Files (Arquivos) menu was developed to perform computer-aided tasks related to text files reading and writing, as well as files compression and decompression. The menu items are shown on Figure 1. a) Item Create ECM_DI.bat (Criar o MCE_DI.bat) This menu item is used to create the text file that optimizes the MC simulation. The produced file comprises data that allow the execution of all internal sources available for ECM MSTA. Each line in the file is an instruction to run via Windows Command Prompt, a ECM with a given file containing the data on the shock sections of the materials that make up the phantom used in the, and another file containing additional data. b) Item Create DataFiles_DI.txt (Criar o ArquivoDados_DI.txt) The simulation generates several text files with specific dosimetry results. The menu item Criar o ArquivoDados_DI.txt, shown on Figure 1, was developed to read such files and write them into a single file called DataFiles_DI.txt (ArquivosDados_DI.txt). The reading and writing mechanisms of the text file in this activity as well as in other activities implemented at DoRadIo are in the classes FileStream, StreamWriter and StreamReader of namespace System.IO, contained in the NET Framework(2) class library. c) Item Compress/Decompress txt Files (Compactar/Descompactar Arquivos txt) As the Data Files_DI.txt (ArquivoDados_DI.txt) is relatively large to be added as a resource to the DoRadIo executable, a tool was created with compression and decompression methods native at namespace System.IO. The main classes used in the implementation of such activities are MemoryStream and DeflateStream, i.e., the data flow is stored in memory during execution. The difference between compressing and decompressing of the text file in this mechanism is informed by one of the two items listed under CompressionMode: compression or decompression. The Digital Images Processing menu (Processamento de Imagens Digitais) Digital Images Processing (Processamento de Imagens Digitais) is the second menu shown in the interface on Figure 1. It comprises two menu items, as follows: one to select and save a given region of interest (ROI) on an image, and another one to count the pixel intensity on the saved images. Such counts per ROI are used to weight dosimetry results when more than one source is used in the simulation. The ratio between the count on a given ROI and the total count on the image is used as a factor by which the dosimetry values for each involved source should be multiplied. Each simulation is always monoenergetic, and the photons emerge from a single source organ. At the beginning of each history simulation, the photon released position and its initial direction of travelling are uniform probability density functions within the parallelepiped containing the source organ, as in the MIRD (Medical Internal Radiation Dose) formulation(7). One of the advantages of performing the monoenergetic simulation and weighting the results is decreasing statistical errors, as all simulations are performed with the same number of histories and the weighting is based on dosimetry results obtained with the plain sources. The pixels' intensity on the hot spots of an 8-bit nuclear medicine image reflects the activity distribution. Thus, by cumulatively counting the intensity on a given ROI, one obtains a number corresponding to the quantity of photons originated from that ROI. As the procedure is repeated, for all ROIs on the image, one obtains quantities which, divided by their sum, become factors that weight the quantities of photons originated from each ROI. The results of the absorbed dose per accumulated activity that are entry data in the DoRadIo software were obtained as if such factors were equal to 1, i.e., each source organ was separately simulated. When there is more than one source organ involved in the simulation, the dosimetry results from the separate simulations of those organs are weighted by those factors. a) Region of interest image acquisition A ROI image is obtained by a logical operation AND involving a mask of defined dimensions over all the image pixels. Each output pixel results from the convolution of pixels defined on the region of the image with the same dimensions of the mask by weights attributed to the operation AND (1 or true). Such an operation was implemented at DoRadIo by reading, at each point (x,y), the pixel intensity of the nuclear medicine image (a) and the corresponding intensity on a mask (b). The mask has the same dimensions as the image, with 0 pixel intensity at all locations except for the desired ROI, which is selected using the mouse. The value returned by the operation is a AND b, i.e., the resulting image contains the mask background and the pixels located in the ROI on the nuclear medicine image(8). b) The menu item Pixel Intensity Count on an Image (Conta Intensidade dos Pixels em uma Imagem) The menu item Pixel Intensity Count on an Image (Conta Intensidade dos Pixels em uma Imagem) has a built-in tool that calculates the sum of the pixels' intensities on an image. For this purpose, a C# function was developed, which receives an image with a single ROI and background filled by 0s (zeros). On a 256-bit vector of integer variables, the frequencies (y) of each gray shade on the image (x) are cumulatively counted. For frequencies different from 0, the function performs the convolution of the gray shade by the frequency. The result is given as the sum of the pixels' intensities of the ROI contained on the entry image. The present study results are presented below. A sequence that presents the main functionalities of the developed software was selected for the case of a hypothetical user interested not only in the dosimetry results, but also in how to prepare the data entry into the ECMs available at DEN-UFPE. RESULTS Most probably, the DoRadIo software functionalities which will raise greater interest from internal dosimetry investigators will be those items related to the obtention of graphical and numerical results. Such users may be interested in analyzing the distribution of the dose originated from a single source organ (plain source) or from a set of organs (weighted sources). The results using plain sources can be obtained with a single click. However, in the case of weighted sources, more actions will be required from the user. In order to demonstrate the potentialities of the application, the steps that must be taken by the user are presented and discussed below, from the preparation of the entry files for ECM MSTA execution to the graphical and numerical results analysis. Preparation of the entry files and Monte Carlo simulation Among the available material on the ECM MSTA material (available at: www.caldose.org), there are 27 text files with data on the source organs most commonly used in the medical practice. Such standard files are prepared to execute a hypothetical 15-energy spectrum: 10, 15, 20, 30, 50, 60, 70, 80, 100, 200, 500, 1000, 1500, 2000, 4000 keV. Such energy interval comprises the photon emitting sources used in medicine. In the case user deals with an intermediate energy, he may interpolate the dosimetry results available in the application. Some adjustments were made in the files with the standard sources. The number of histories was established at 108, independently of the energy and of the source organ; and the names of the exit files were customized. When clicking on the item Create the ECM_DI.bat (Criar o MCE_DI.bat) of the Files (Arquivos) menu, the user can create a text file, the ECM_DI.bat, with data to execute the 27 plain sources. The simulation was triggered and took approximately 30 continuous days on a PC at LDN-IFPE. In this case each source were separately simulated, 27 user inputs would be necessary. Once the process was completed, 405 files (15 energies × 27 sources) with specific dosimetry results were obtained. Data organization and compression As the Create DataFiles_DI.txt (Criar ArquivosDados_ DI.txt) item of the Files (Arquivos) menu is triggered, the software reads the 405 files with specific dosimetry results obtained as described in the previous item, and organizes them into a single file called DataFiles_DI.txt (Arquivos Dados_DI.txt). Such file is used to obtain dosimetry results at a considered target: one source (Plain Sources) or N sources (Weighted Sources). The file DataFiles_DI.txt (ArquivosDados_DI.txt) is relatively large (27,066 kB) to be added as a resource of the DoRadIo. By the Compress/Decompress txt Files (Compactar/Descompactar Arquivos txt) item of the Files (Arquivos) menu, the file was compressed to 8,721 kB, reducing the storage space by 67.8%. The compressed file was added as a resource and it is read and decompressed during execution by the software interfaces to present specific results. Obtaining factors for a Weighted Source Nuclear medicine images may be used to estimate the weight of source organs in a simulation. In the presented example, one used the image of an adult female patient (Figure 2) obtained from a catalogue constructed as a reference(9). The ROIs were defined on the image based on the pixels intensities. On Figure 2, it is possible to observe three ROIs: a more intense one in the neck region (thyroid), another in one the bowel loop region (small bowel) and a third one in the hip region (bladder). 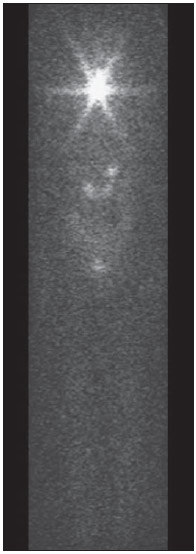 Figure 2. Nuclear medicine image obtained from a catalogue(9). The application of the AND tool is demonstrated below, on a step-by-step basis (Figure 2). Step 1: The interface shown on Figure 3 is loaded by clicking on the menu item that performs the AND operation. Step 2: An image is opened by clicking on (1) the interface. In the frame on the left, the image shown on Figure 2 is adjusted. Step 3: The first ROI is selected (neck region) with a click on the left button of the mouse to select the upper left corner of the ROI, and another click on the right button to select the lower right corner in such a way to form a rectangle (2). The button (3) must be clicked in order to generate and/or update the images from the mask and from the ROI (4). Step 4: Finally, one clicks on it (5) in order to save the images of the mask and of the ROI. Step 5: Steps 1 to 4 are repeated to save other ROI images. On a single image, Figure 4 presents the results obtained with the application of the tool in the three regions with the respective saved images of the ROIs. The image on Figure 4 contains 0s (zeros) as background and gray shades in the intervals [0, 255] at the ROIs. By using the item Image Pixels Intensity Count (Conta Intensidade de Pixels de uma Imagem) in the Digital Image Processing (Processamento de Imagens Digitais) menu one obtained a total of 3042, 1271 and 432 for the sum of pixels intensities for the thyroid, small bowel and bladder, respectively. Such values divided by their total sum were utilized as weighting factors for such sources.  Figure 4. Image of the ROIs selected on Figure 2. In order to obtain a realistic distribution of doses from photons coming from the three sources, such a count is indispensable, as the total intensities in the ROIs are proportional to the quantity of photons whose histories initiate at such regions. Thus, the ratios between the total intensities for each ROI, shown on Figure 4, and the total on the image are, respectively, the normalized weights of the simulated sources 0.641, 0.268 and 0.091. Such weights are applied on the dosimetry data built in the software and the user can save the results in an external file. Dosimetry results presentation and analysis Figure 5 shows four menu items, where only ECM MSTA is enabled in the current release of DoRadIo. The items in this menu allow the display of dosimetry results from plain sources and the creation and display results from weighted sources. 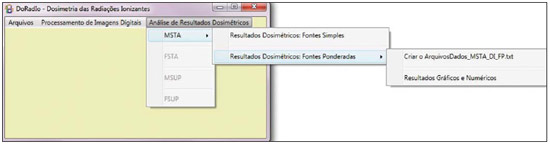 Figure 5. The Dosimetry Results Analysis (Análise de Resultados Dosimétricos) menu and its sub-menus. a) Dosimetry results utilizing Plain Sources As the sub-item Dosimetry Results: Plain Sources (Resultados Dosimétricos: Fontes Simples) is selected (Figure 5), the user can obtain numerical and graphical results of the action from a given source organ on a determined target organ. For example, Figure 6 shows the loaded UI when the organ source (THYROID) and a target organ (LIVER) were selected and the button Dosimetry Results Tables (Tabelas com Resultados Dosimétricos) was clicked on. With that action the table with the values of the following conversion coefficients for 15 energies, was loaded: Accumulated Dose/Activity, Emitted Dose/Particle, Absorbed Fraction and Specific Absorbed Fraction. The statistical error evaluated by the coefficient of variance function(10), is also shown on the table. In the box Select a Chart Option (Escolha uma Opção de Gráfico), the option Accumulated Dose/Activity (mGy/MBq.s) [Dose/Atividade Acumulada (mGy/MBq.s)] was selected. As a result, two charts are shown: Accumulated Dose/Activity (mGy/MBq.s) × Energy (keV) [Dose/Atividade Acumulada (mGy/MBq.s) × Energia (keV)] and Variance Coefficient (%) × Energy (keV) [Coeficiente de variância (%) × Energia (keV)]. 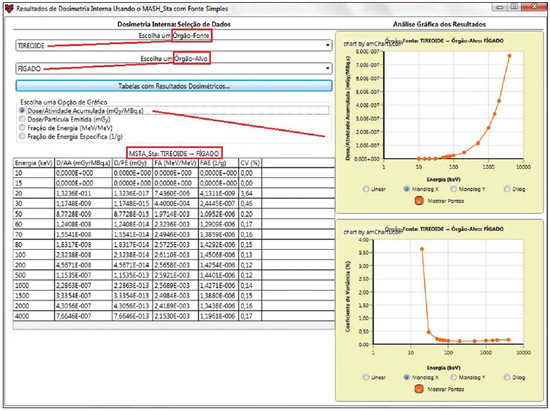 Figure 6. Window resulting from the Dosimetry Results: Plain Source (Resultados Dosimétricos: Fonte Simples) menu, with an example of the user choices. b) Dosimetry results utilizing Weighted Sources The sub-item menu Dosimetry Results: Weighted Sources (Resultados Dosimétricos: Fontes Ponderadas) allows the user to create a file with the results from the action of more than one source organ on a target organ and to analyze them. As shown on Figure 5, such sub-item comprises two other items: Create DataFiles_MSTA_DI_FP.txt (Criar o ArquivoDados_MSTA_DI_FP.txt) and Numerical and Graphical Results (Resultados Gráficos e Numéricos). When the item Create DataFiles_MSTA_DI_FP.txt (Criar o ArquivoDados_MSTA_DI_FP.txt) was selected, the UI on Figure 7 was loaded, and the normalized weights obtained in the item Obtaining factors for a Weighted Source were entered. By clicking on the button Save DataFiles_MSTA_DI_FP.txt (Salvar ArquivosDados_ MSTA_DI_FP.txt) one saves a text file with the name of the caption. Such file contains the weighted dosimetry results for all available targets at MSTA and is used in the numerical and graphical analysis.  Figure 7. Interface for definition of source organs with their respective weights. By clicking on the item Numerical and Graphical Results (Resultados Gráficos e Numéricos) shown on Figure 5, the software loads the interface on Figure 8, where it is possible to select a target organ (for example, the liver). By clicking on the button Dosimetry Results Table (Tabela com Resultados Dosimétricos) the table is shown with the conversion coefficients and coefficients of variance for the selected target organ. In the case user wishes to analyze the coefficient as a function of the energy in keV, he must select the corresponding option. 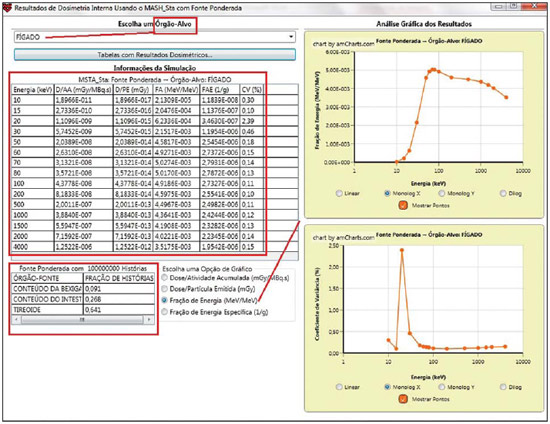 Figure 8. Interface to obtain graphical and numerical results with the weighted source acting on a target organ. DISCUSSION In the present study, the MC simulations were performed with source algorithms based on nuclear medicine images, using the voxel phantom MASH combined with the EGSnrc. In a great number of cases, the images from patients presented more than one hot-spot. Therefore, estimates with weighted sources are most frequently used. In a previous study(9), the fractions were applied to the number of histories during the simulation, which resulted in significant statistical errors caused by the fractioning of such a number. For example, for the fractions obtained in the present study, the simulation would be made for the three sources separately. Thus the total of 108 photons should be weighted between sources, with 6.41 × 107 in the thyroid, 2.68 × 107 in the small bowel, and 9.1 × 106 in the bladder. In the present study, all simulations were performed with 108 histories. The weighting was applied to the dosimetry results. In a future study the accuracy of this approach will be demonstrated with the undertaking of separate simulations for thyroid, small bowel and bladder with the respective history numbers being drawn during the execution time. The comparative analysis will be incorporated into DoRadIo. System errors originated from the ECMs used by GDN/CNPq, which may compromise the accuracy of dosimetry results are constantly being detected and corrected or minimized. An important point that can be improved in the DoRadIo is the determination of the ROIs on nuclear medicine images. Besides the mouse resources, the user could evaluate a threshold gray shade for the contour of the ROIs and the software would, automatically, load as a standard the ROIs limited by such threshold. Subsequent adjustments would be made by the user. CONCLUSIONS The present study presents the first version of the DoRadIo software, which provides:
The computer-aided tools developed by GDN are free and are available at http://dosimetrianumerica.org/. Additionally, such tools are under continuous improvement. REFERENCES 1. Leal Neto V. SID: um sistema computacional para obtenção e gerenciamento de informações sobre dosimetria das radiações ionizantes. [Tese de doutorado]. Recife: Universidade Federal de Pernambuco; 2013. 2. Sharp J. Visual C# 2010 step by step. 1ª ed. Washington: Microsoft Press; 2010. 3. Vieira JW, Leal Neto V, Lima Filho JM, et al. Transformation of the normal distribution for Monte Carlo modeling of regions of adult trabecular bones for use in computational models of exposure. 13th International Congress of International Radiation Protection Association; 2012 May 13-18; Glasgow, Scotland. 4. Vieira JW, Lima FRA. A software to digital image processing to be used in the voxel phantom development. Cell Mol Biol (Noisy-legrand). 2009;55:16-22. 5. Kawrakow I, Rogers DWO, Mainegra-Hing E, et al. The EGSnrc Code System: Monte Carlo simulation of electron and photon transport. Report No. PIRS-701. Ottawa, Canada: National Research Council Canada; 2011. 6. Cassola VF. Desenvolvimento de fantomas humanos computacionais usando malhas poligonais em função da postura, massa e altura. [Tese de doutorado]. Recife: Universidade Federal de Pernambuco; 2011. 7. Zaidi H, Sgouros G. Therapeutic applications of Monte Carlo calculations in nuclear medicine. 1ª ed. Bristol: IOP Publishing; 2003. 8. Gonzalez RC, Woods RE. Digital image processing. 3ª ed. New Jersey: Prentice Hall; 2008. 9. Lopes Filho FJ. Avaliações dosimétricas em pacientes submetidos à radioiodoterapia com base em fantomas de voxels e em imagens de medicina nuclear. [Tese de doutorado]. Recife: Universidade Federal de Pernambuco; 2007. 10. Santoro A, Oguri V, Mahon JR, et al. Estimativas e erros em experimentos de física. 2ª ed. Rio de Janeiro: UERJ; 2008. 1. PhD, Teacher and Researcher at Instituto Federal de Educação, Ciência e Tecnologia de Pernambuco (IFPE), Recife, PE, Brazil 2. PhD, Researcher, Member of the Department of Nuclear Energy Post-graduation Program - Universidade Federal de Pernambuco (UFPE), Teacher at Escola Politécnica de Pernambuco - Universidade de Pernambuco (UPE), Recife, PE, Brazil 3. PhD, Researcher, Member of the Department of Nuclear Energy Post-graduation Program - Universidade Federal de Pernambuco (UFPE), Researcher at Centro Regional de Ciências Nucleares (CRCN), Recife, PE, Brazil. Mailing Address: Dr. Viriato Leal Neto Instituto Federal de Educação, Ciência e Tecnologia de Pernambuco Avenida Professor Luís Freire, 500, Cidade Universitária Recife, PE, Brazil, 50740-540 E-mail: viriatoleal@yahoo.com.br Received March 13, 2013. Accepted after revision October 28, 2013. Study developed at Department of Nuclear Energy - Universidade Federal de Pernambuco (UFPE), Recife, PE, Brazil. |
|
Av. Paulista, 37 - 7° andar - Conj. 71 - CEP 01311-902 - São Paulo - SP - Brazil - Phone: (11) 3372-4544 - Fax: (11) 3372-4554